By Jessie Lau, Biochemistry and Molecular Biology ‘20
Authors Note: Since the advent of Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR Associated Protein 9 (CRISPR/Cas9) discovery and biotechnological breakthroughs thereafter, this revolutionary application has been primarily focused on human health, particularly fostering solutions to numerous debilitating ailments. However, the general public has offered little attention towards the use of this engineering feat in a broader ecological system. Upon watching the new Netflix original Unnatural Selection, discussion of considering the use of CRISPR/Cas9 in New Zealand’s effort to completely eradicate invasive species piqued my interest. The following article is an exploration of CRISPR/Cas9 prospect into New Zealand’s bold environmental pursuit and its potential ecological impact.
Abstract
Since it has become feasible to cross oceans to reach unforeseeable land masses, invasive alien species (IAS) are an increasing threat to international biodiversity. Moreover, no other region faces as great of a peril as New Zealand (NZ), which holds the record as the nation with the highest survival rate of threatened avifauna (birds of a particular region) species [5]. In 2012, New Zealand physicist Sir Paul Callaghan introduced a large-scale eradication program to permanently remove eight invasive mammalian predators (rodents: Rattus rattus, Rattus norvegicus, Rattus exulans, Mus musculus; mustelids: Mustela furo, Mustela erminea, Mustela nivalis; and the common brushtail possum: Trichosurus vulpecula) [5]. Four years after this grand proposal, the NZ government committed to a national challenge titled “Predator Free 2050” (PF 2050) to pursue this audacious goal.
Introduction
Approximately 85 million years ago, NZ was one of the first landmasses to split from the supercontinent Gondwana and as it shifted away, it did not carry mammals until bats flew and aquatic species swam to this island. [7]. With the introduction of rodent species initially through Polynesian settlement some 750 years ago and thereafter European seafaring ventures, the endemic species of NZ have been left vulnerable to novel predators. Consequently, at least 51 native bird species that have evolved adaptive skills of remaining close to the ground have been unsuccessful at surviving alongside these rodent predator species [4]. These invasive omnivorous rodents prey on birds, eggs, seeds, snails, lizards, and fruits. As a result, their varied diets prompt competition with the native fauna, further placing pressure on their vulnerable survival [7]. Despite CRISPR/Cas9 modifications offering the greatest potential in gene drive to eliminate these unwanted predators, several techniques implemented to control these invasive populations, such as pesticides, trapping innovations, and biological factors have given favorable results.
Past Successes and Future Endeavors
For decades, the NZ government has pursued eradication initiatives to permanently eliminate foreign invasive species on small local islands. Through concerted efforts, several of these projects have proven successful in the restoration of the natural biodiversity this island nation boasts.
In June of 2009, the NZ Department of Conservation (DOC) undertook a multi-action plan to simultaneously eradicate rodent, rabbit, stout, hedgehog, and cat species in Rangitoto and Motutapu islands. After three years of aerial dispersion of anticoagulant Pestoff 20RTM combined with trapping and indicator dogs, the island witnessed total elimination of the islands’ stoats and four rodent species; declination of rabbit and hedgehog population by 96%; and over 50% reduction in cats [7].
Despite these successful feats, the invasive species fecundity and ability to adapt to these challenges still present an overwhelming challenge to reach the goal of complete eradication. Recently, more direct approaches delving into novel genetic inheritance techniques have been explored to serve as a potential permanent solution. Termed the“Trojan Female Technique” (TFT), the method makes use of the correlation of sperm fitness dependence on the abundance of mitochondrial DNA (mtDNA) [2]. Healthy sperm is dependent on sufficient mitochondrial level for energy production to manage motility and fertilization. For example, experiments conducted on Drosophila melanogaster supports the causation of reduced spermatogenesis and sperm maturation due to induced mutations in cytochrome mitochondrial gene [9]. Contrasting female eggs, the asymmetric greater dependence of sperm on mtDNA for normal functionality results in only male populations to be the sole source of target. Induction of mitochondrial mutations in females to compromise total sperm viability in future male progenies will serve as an effective control to population growth.
Although seemingly promising, TFT is not guaranteed to completely eradicate propagation for several reasons. For starters, males with impaired fertility can still provide sufficient sperm count to fertilize eggs on a population-based scale. Furthermore, in circumstances where females do receive nonviable sperm count, they can still seek adequate functioning sperm through matings with other males. On a larger scale, should the mutation pervade, selection pressures could still inadvertently choose for nuclear modifications to make up for the mitochondrial defects [2]. These flaws raise the need for more pervasive and permanent resolutions.
Daisy Chain CRISPR Gene Drive
The recent biochemical breakthrough underpinning the ability to effectively and precisely modify genes with CRISPR/Cas9 has allowed for potential biotechnology to boom in the realm of ecology. The simple generation of a short RNA sequence into a virus or bacterium to serve as a vector, guides the cutting mechanism of Cas9 to specific regions in the genome to be excised, prompting for these double stranded breaks to be fixed through DNA repair mechanisms. While these fixtures can potentially repair the gene, it can also raise the possibility for the gene to be disabled, introduce a new function, or create an unforeseen mutation. Given the right specific targeting in the germ line, this approach houses the innovation for exterminating entire species through gene drive [6].
The mechanism behind gene drive overthrows the traditional Mendelian sexual reproduction concept of proportional contribution from both the male and female parent. The power comes from the ability of one genetically modified (GM) contributor encoding for the ‘gene drive’ to cut the other set of chromosomes lacking these genes and replace this excision with a self-replicating copy. In effect, this divisive modification would push for an otherwise heterozygous offspring from a wild-type mating with a GM partner to become homozygous for certain genes to be carried on and propagated by future generations. Given that these genetic alterations do not affect the fitness of the organism, dissemination of 1% of the population with CRISPR modified genes can lead to 99% of the local population carrying the genetic indicator in as little as nine generations (The use of gene editing to create gene drives for pest control in New Zealand).

Such a unilateral approach poses political and ethical challenges amongst neighboring nations with diverging ecological approaches to confront pest control. Should this pervasive gene drive program reach beyond its intended border, great difficulty would arise in maintaining this ecological enclosure. For example, possum is on the list of invasive species in NZ while just 2,500 miles west, its neighbor Australia keeps this species of marsupials under protection. As such, scientists have devised a simple model to localize gene alterations, coined The Daisy Chain.
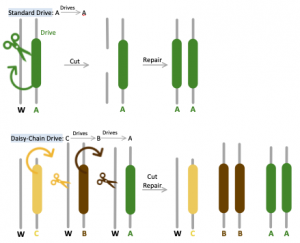
Unlike the original gene drive method in which all components necessary for transformation (CRISPR, edited DNA, and guide RNAs) are provided on the same chromosome, Daisy Chain provides a self-exhaustive means of guaranteeing genetic edits. This tool is designed so that each component required for genetic alteration is dependent on the presence of a different element upstream on the gene found on the same locus to be activated [8]. The most downstream portion of this chain contains the “load” of engineered dominant lethal genes preventing reproduction, which will be promoted to higher frequency in the population within several generations. For instance, should an engineered allele contain three elements A, B, and C, element C would render element B to drive, which will in turn cause element A carrying the final load to drive. The initial element (C in this case) does not actually drive, thus is restricted by the number of altered individuals released into the wild and will be lost via natural selection over time. During initial implementation, the presence of C will increase B in abundance, but B will eventually decline and finally disappear as C is lost in the population. The rapid rise in abundance of B will also cause A to increase in frequency within the local population; however, with the decrease of B, A would not be driven and will ultimately vanish as well [1]. Using MIT Professor Esvelt’s analogy, “… the elements of a daisy drive system are similar to booster stages of a genetic rocket: those at the bottom of the base of the daisy-chain help life the payload until they run out of fuel and are successively lost.”
Challenges with Daisy Chain CRISPR Gene Drive
CRISPR/Cas9 technology’s ability to potentially alter these invasive species’ fecundity provides an avenue of pursuing NZ’s goal of PF 2050. Despite the developed understanding of how to carry out this plan, scientists in NZ are still working to piece together the genomes of stoats and possums in order to understand where to properly facilitate the engineered RNA sequence. Other barriers that must be acknowledged are the unprecedented approach to genetically modify marsupials and the difficulty of implanting hundreds to thousands of oocytes to be dispersed amongst their population.
Beyond these known difficulties, scientists still tread in unknown terrains pertaining to whether these mutations can have pernicious effects in the survival, health, and reproductive success in propagating these mutations within their populations. Further exploration into the development of these modifications, and the potential impacts they can have on these animals, must be investigated on model organisms prior to widespread use.
Of the eight listed mammalian species vied to be permanently eradicated from NZ, Mus musculus holds the most promise, given the extensive knowledge of the Mus musculus genome. With the help of scientists outside of New Zealand, joining in on the efforts to identify which germline gene to focus on, this project has received international attention.
Although the daisy drive provides promising potential, research collaborators at MIT and Harvard have identified a possible risk of, “… DNA encoding a drive component from one element to another, thereby creating a ‘daisy necklace’ capable of a global drive” [1]. Due to this rare recombinatory event arising from the similarity of DNA sequences, these investigators have looked into circumventing the problem by creating numerous alternatives to CRISPR components and selecting the model with the greatest diversity.
Conclusion
From their renowned aviary to reptilian species, New Zealand’s islandic geographical region houses some of the most biodiverse fauna known to man. The arrival of human settlement has introduced predatory species, causing endemic species to experience extinction at concerning rates [4]. With the purpose of preserving their unique remaining diversity, New Zealand has committed to concerted efforts of varying methods to eradicate these invasive vertebrate pests. Investigation into genetic modifications can provide for more expansive and thorough techniques to eliminate these human introduced pests and allow for these endangered species to thrive once again. By further exploring daisy chain CRISPR/Cas9, this effort can be genetically inherited by offspring, allowing for nature to carry out this effort. As opposed to continued efforts of targeting each individual one by one, conservation ecologists can borrow from molecular biologist’s toolkit to revolutionize the means of pursuing pest control and perhaps even pave the road for future endeavors with similar pursuits.
References
- Esvelt, Kevin M. “Daisy Drives.” Sculpting Evolution, www.sculptingevolution.org/daisydrives.
- Gemmell, Neil J., et al. “The Trojan Female Technique: a Novel, Effective and Humane Approach for Pest Population Control.” Proceedings of the Royal Society B: Biological Sciences, vol. 280, no. 1773, 2013, pp. 1–6., doi:10.1098/rspb.2013.2549.
- Griffiths, Richard, et al. “Successful Eradication of Invasive Vertebrates on Rangitoto and Motutapu Islands, New Zealand.” Biological Invasions, vol. 17, no. 5, 2014, pp. 1355–1369., doi:10.1007/s10530-014-0798-7.
- Owens, Brian. “The Big Cull: Can New Zealand Pull off an Audacious Plan to Get Rid of Invasive Predators by 2050?” Nature, vol. 541, 12 Jan. 2017, pp. 148–150.
- Russell, James C., John G. Innes, Philip H. Brown, and Andrea E. Byrom. “Predator-Free New Zealand: Conservation Country.” BioScience 65, no. 5 (October 2015): 520–25. https://doi.org/10.1093/biosci/biv012.
- Saey, Tina Hesman. “Explainer: How CRISPR Works.” Science News for Students, 4 Dec. 2017, www.sciencenewsforstudents.org/article/explainer-how-crispr-works.
- “Why Predator Free 2050?” Department of Conservation. Accessed November 18, 2019. http://www.doc.govt.nz/nature/pests-and-threats/predator-free-2050/why-predator-free-2050/.
- Dearden, Peter K., et al. “The Potential for the Use of Gene Drives for Pest Control in New Zealand: a Perspective.” Journal of the Royal Society of New Zealand, vol. 48, no. 4, 2017, pp. 225–244., doi:10.1080/03036758.2017.1385030.
- Wolff, Jonci N., et al. “Mitonuclear Interactions, MtDNA-Mediated Thermal Plasticity and Implications for the Trojan Female Technique for Pest Control.” Scientific Reports, vol. 6, no. 1, 2016, doi:10.1038/srep30016.
- Min, John, Jason Olejarz, Joanna Buchthal, Alejandro Chavez, Andrea L. Smidler, Erika A. DeBenedictis, George M. Church, Martin A. Nowak, Kevin M. Esvelt, and Charleston Noble. “Daisy-Chain Gene Drives for the Alteration of Local Populations.” PNAS. National Academy of Sciences, April 23, 2019. https://www.pnas.org/content/116/17/8275.